Medical AI is about to become more powerful and versatile thanks to the fast creation of reusable AI models that can learn from different types of data. We call this new approach a foundation model for medical AI (FMAI). FMAI models will be able to do many different tasks with little or no data that is labeled for each task. They will learn by themselves from large and varied datasets, and they will be able to handle different kinds of medical data, such as images, records, tests, genes, networks, or text. They will also be able to generate rich outputs, such as written summaries, spoken advice, or image annotations that show advanced medical thinking skills.
In this project, we explore how to construct such FMAI models and apply them to the diversity of medical settings. Research questions include:
(i) Construct large-scale FMAI models trained on multi-modal data using self-supervised or semi-supervised algorithms. The algorithm can learn with heterogeneous, mixing data and capture hidden semantic relationships among domains.
(ii) how to efficiently fine-tune FMAI models to a new task using very little labeled sample or ideally without the need for new annotated data (zero-shot learning) [1, 2].
(iii) constructing FMAI models so that they can accept inputs and produce outputs using varying combinations of data modalities (for example, images, text, test results, or any mix of them). This flexible mechanism will allow users to interact with models by customizing queries, making insights easier, and offering more freedom across medical tasks.
(iv) Represent medical knowledge into FMAI so that they can reason predictions on previously unseen tasks and use medically accurate language to explain their outputs.
Our recent works toward these aims include novel intra-inter clustering-based self-supervised algorithms for medical imaging [4], and lvm-med [5], the first large-vision model for medical imaging trained on millions of images with diverse data molarity and body organs.
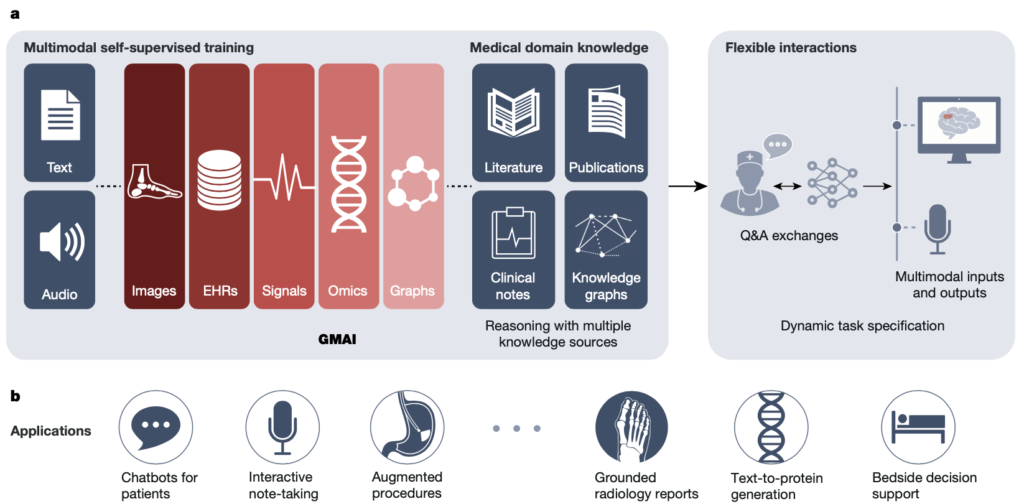
Figure 1. (a) data type can be used to train the FMAI model, (b) applications of FMAI models in medical settings. Source [3]
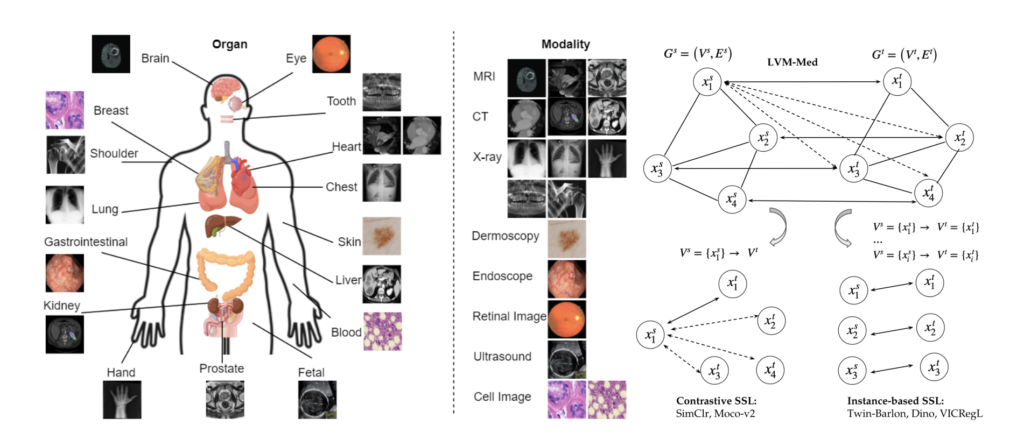
Figure 2. LMV-Med algorithm. (left) Overview of the body organs and modalities in our collected dataset; (right) LVM-Med unifies and extends contrastive and instance-based self-supervised learning approaches by specifying the graph’s properties.
References
[1] Alayrac, Jean-Baptiste, et al. “Flamingo: a visual language model for few-shot learning.” Advances in Neural Information Processing Systems 35, 2022.
[2] Brown, Tom, et al. “Language models are few-shot learners.” Advances in neural information processing systems 33, 2020.
[3] Moor, Michael, et al. “Foundation models for generalist medical artificial intelligence.” Nature, 2023.
[4] Nguyen, Duy MH, et al. “Joint self-supervised image-volume representation learning with intra-inter contrastive clustering.” Proceedings of the AAAI Conference on Artificial Intelligence, 2023.
[5] Nguyen, Duy MH, et al. “LVM-Med: Learning Large-Scale Self-Supervised Vision Models for Medical Imaging via Second-order Graph Matching.”, Arxiv Preprint 2023.