Ophthalmo-AI
Age-related macular degeneration (AMD) is the primary cause of severe vision loss in the western world. One cause is the retinal accumulation of harmful byproducts of metabolism processes occurring in the eye. Treatment aims at slowing down disease progression and is performed through the injection of anti-VEGF agents into the eye. These injections are administered according to the current state of the disease, for which the patient is monitored within regular intervals. One method to evaluate disease progression is the assessment of Optical Coherence Tomography (OCT) images. These images show retinal layers and other medical markers in a pseudo-histological fashion. This technique uses the reflection of near-infrared light to create a 3D-representation of the retina, using the intensity of the reflection at different depths and from different tissue types. With the help of OCT images, the ophthalmologists can observe the state of the individual layers and the presence of sub- or intraretinal fluid that is often present if the disease is active.
Ophthalmo-AI is a research project supported by the Bundesministerium für Bildung und Forschung. The project’s research focuses on active learning (AL) and decision support systems using AI in the medical field (1). We use active learning to counter the sparsity of annotated data in the medical field. Especially deep learning (DL) systems rely on large sets of annotated data. Obtaining annotations in the medical domain is expensive, since often only medical experts are qualified to provide those. While most of the existing computer vision literature is centered on how to use AL for classification tasks, we conduct experiments to evaluate AL for medical image segmentation.
Active Learning for OCT Segmentation
Active learning is a paradigm in supervised machine learning (ML) where the model interacts with a user to label new data points. It is often used in scenarios with a large pool of unlabeled data where the labeling process is expensive. By using the ML model to select which examples to learn from, the algorithm can learn a concept with fewer examples than traditional supervised learning. This work focuses on pool-based AL, i.e., uses a fixed data pool to select samples for annotation. Active learning is an iterative process that starts with a small, annotated data set to train the ML model. This model is then used to query the remaining data from the unlabeled pool. Querying refers to the process of assigning scores to individual data points, according to how informative they are. The most informative data points are then selected for the next annotation round and added to the training data set. The cycle is repeated until the model reaches a given target performance.
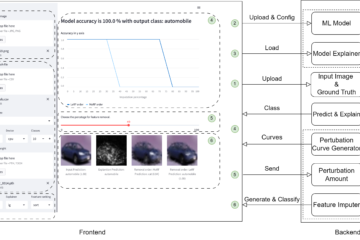
To support our research with AL we build MedDeepCyleAL, an end-to-end framework implementing the complete AL cycle, allowing researchers the flexibility of freely choosing the type of deep learning model they want to employ, and which provides an annotation tool (AT) that supports classification and segmentation of medical images. Previous projects in this area do not include an end-to-end solution, do not focus on deep learning, or must be integrated programmatically. It optimizes the annotation and training cycle of DL models in AL. MedDeepCyleAL consists of four components: an annotation tool for medical image segmentation and classification, a controller for executing individual AL iterations, data management for continuous updates of the data set throughout iterations, and the core AL backend that performs the training of the DL model. This platform enables us to easily research different aspects of AL.
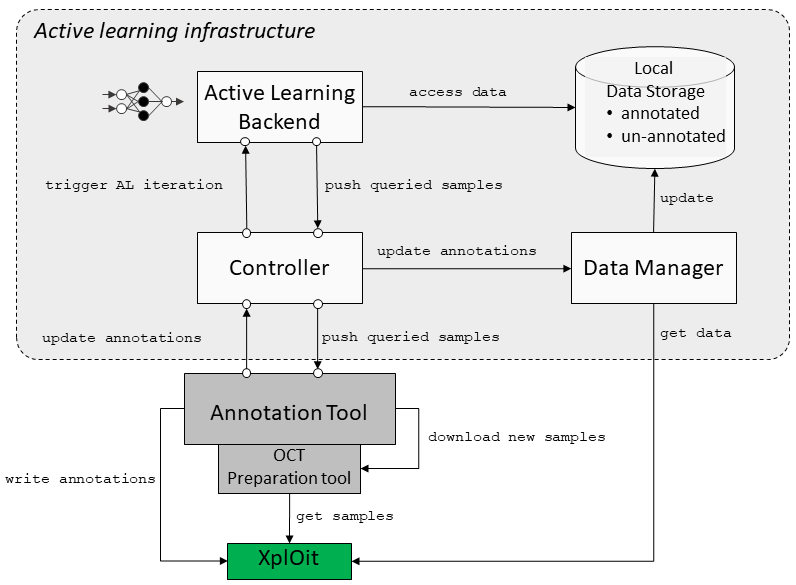
Figure 1: Active Learning Architecture
One research topic is partial labelling. We refer with this term to the process of only annotating parts of an image (only specific layers) in or. We hypothesize that some parts of the retinal images from AMD patients are easier to segment than others. While the upper-most layer is easily distinguished from the background/vitreous eye, distinguishing neighboring layers is more challenging. We are training a DL model on retinal OCT images, whose input is a 2D image of the retina and that creates a (semantic) segmentation of the input image. While the common approach involves training a model on a fully labeled image, our approach only requires labeling in specific areas. Which areas will be presented to the annotator for labeling is determined through AL. Note that in most AL approaches, the AL algorithm would be used to compile a training set for a deep learning model. In our experiment, we use a pre-defined, fixed set of images. With the help of AL, it is determined what layers/medical markers of an individual image will be annotated. Instead of training the model with a fully annotated ground truth, we use a weighted loss that effectively masks out non-annotated areas. Thus, we reduce annotation cost not by reducing the required data set size to attain a certain score but by reducing the annotation effort needed for individual images.
Along with active learning, another paradigm to reduce the annotation effort is self-supervised training that learns from a large pool of unlabeled data in an unsupervised way. Self-supervised learning (SSL) has gained attention as it can utilize large amounts of unlabeled data, which are often readily available. It leverages the inherent structure or information present in the input data itself to create supervised learning tasks. The model learns to predict certain aspects of the data, such as filling in missing parts, generating transformed versions, or clustering groups of similar samples closer and diverse samples afar (contrastive learning). Recent studies suggest that using self-supervised learning with active learning can be a powerful combination that leverages the benefits of both approaches. Neural models for our task can be pre-trained using large pools of unlabeled OCT slices by applying self-supervised tasks in order to learn meaningful representations of the data. By integrating SSL optimization in the sample selection of our active learning framework, we plan to evaluate their performance in the medical imaging domain.
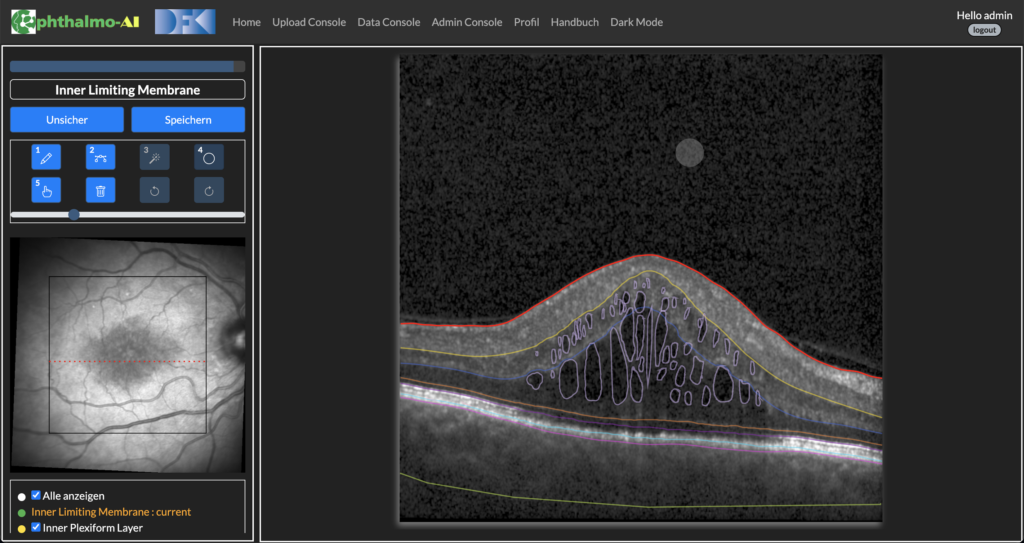
Figure 2: Annotation Tool
References
- Kadir, Md Abdul, Hasan Md Tusfiqur Alam, and Daniel Sonntag. “EdgeAL: An Edge Estimation Based Active Learning Approach for OCT Segmentation.” arXiv preprint arXiv:2307.10745 (2023).