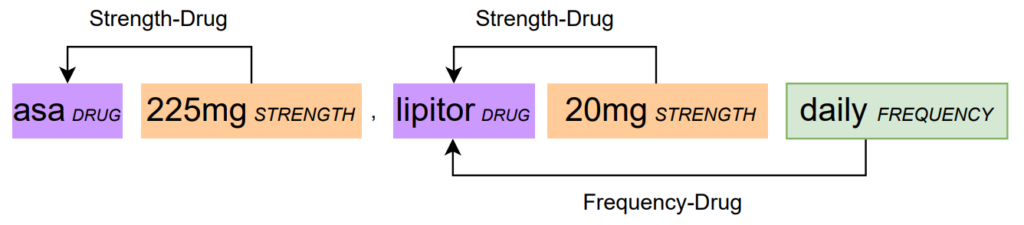
Example from n2c2 of relation extraction [1]
This work explores the effectiveness of employing Clinical BERT for Relation Extraction (RE) tasks in medical texts within an Active Learning (AL) framework. Our main objective is to optimize RE in medical texts through AL while examining the trade-offs between performance and computation time, comparing it with alternative methods like Random Forest and BiLSTM networks. Comparisons extend to feature engineering requirements, performance metrics, and considerations of annotation costs, including AL step times and annotation rates. The utilization of AL strategies aligns with our broader goal of enhancing the efficiency of relation classification models, particularly when dealing with the challenges of annotating complex medical texts in a Human-in-the-Loop (HITL) setting.
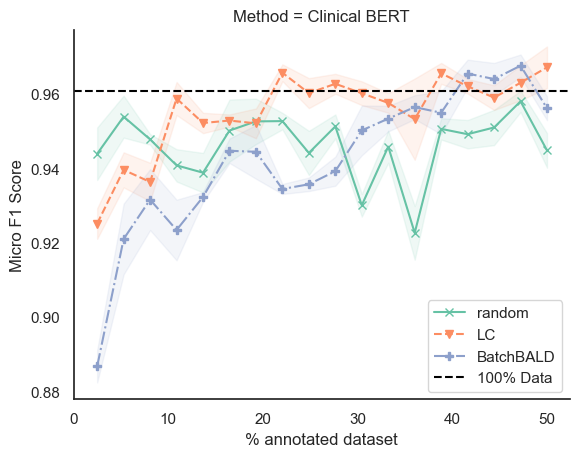
The results indicate that uncertainty-based sampling-Least Confidence (LC) achieves comparable performance with significantly fewer annotated samples across three categories of supervised learning methods, thereby reducing annotation costs for clinical and biomedical corpora. While Clinical BERT exhibits clear performance advantages, the trade-off involves longer computation times in interactive annotation processes. In real-world applications, where practical feasibility and timely results are crucial, optimizing this trade-off becomes imperative [3].
References
[1] Henry, S., Buchan, K., Filannino, M., Stubbs, A. and Uzuner, O., 2020. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. Journal of the American Medical Informatics Association, 27(1), pp.3-12.
[2] Liang, S., Hartmann, M. and Sonntag, D., 2023. Cross-lingual German Biomedical Information Extraction: from Zero-shot to Human-in-the-Loop. arXiv preprint arXiv:2301.09908.
[3] Liang, S., Sánchez, P.V. and Sonntag, D., 2024, March. Optimizing Relation Extraction in Medical Texts through Active Learning: A Comparative Analysis of Trade-offs. In Workshop on Uncertainty-Aware NLP (UncertaiNLP 2024) (p. 23).